-Search query
-Search result
Showing all 45 items for (author: kirk & ns)

EMDB-18207: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide, Glacios map
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-18230: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA
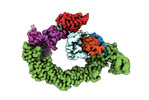
EMDB-18915: 
Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

PDB-8q7r: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

PDB-8r5h: 
Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-42804: 
Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

EMDB-42806: 
Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

EMDB-42807: 
Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

EMDB-18586: 
Cryo-EM reconstruction of crosslinked native Bacillus subtilis collided disome binding MutS2 SMR and KOW domains
Method: single particle / : Park E, Mackens-Kiani T, Berhane R, Esser H, Erdenebat C, Burroughs AM, Berninghausen O, Aravind L, Beckmann R, Green R, Buskirk AR

EMDB-41138: 
CryoEM structure of MFRV-VILP bound to IGF1Rzip
Method: single particle / : Kirk NS

EMDB-18585: 
Cryo-EM reconstruction of Bacillus subtilis collided disome binding MutS2 SMR and KOW domains
Method: single particle / : Park E, Mackens-Kiani T, Berhane R, Esser H, Erdenebat C, Burroughs AM, Berninghausen O, Aravind L, Beckmann R, Green R, Buskirk AR

EMDB-18901: 
Bacillus subtilis MutS2-collided disome complex (stalled 70S)
Method: single particle / : Park E, Mackens-Kiani T, Berhane R, Esser H, Erdenebat C, Burroughs AM, Berninghausen O, Aravind L, Beckmann R, Green R, Buskirk AR

PDB-8r55: 
Bacillus subtilis MutS2-collided disome complex (collided 70S)
Method: single particle / : Park E, Mackens-Kiani T, Berhane R, Esser H, Erdenebat C, Burroughs AM, Berninghausen O, Aravind L, Beckmann R, Green R, Buskirk AR

EMDB-18558: 
Bacillus subtilis MutS2-collided disome complex (stalled 70S)
Method: single particle / : Park E, Mackens-Kiani T, Berhane R, Esser H, Erdenebat C, Burroughs AM, Berninghausen O, Aravind L, Beckmann R, Green R, Buskirk AR

PDB-8qpp: 
Bacillus subtilis MutS2-collided disome complex (stalled 70S)
Method: single particle / : Park E, Mackens-Kiani T, Berhane R, Esser H, Erdenebat C, Burroughs AM, Berninghausen O, Aravind L, Beckmann R, Green R, Buskirk AR
EMDB-29101: 
EGFR:Degrader:VHL:Elongin-B/C:Cul2
Method: single particle / : Rosenberg SC

EMDB-26306: 
Single-chain LCDV-1 viral insulin-like peptide bound to IGF-1R ectodomain, leucine-zippered form
Method: single particle / : Kirk NS, Lawrence MC

EMDB-26363: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM459
Method: single particle / : Kirk NS, Lawrence MC

EMDB-26364: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM462
Method: single particle / : Kirk NS, Lawrence MC

EMDB-24791: 
Head region of a complex of IGF-I with the ectodomain of a hybrid insulin receptor / type 1 insulin-like growth factor receptor
Method: single particle / : Xu Y, Lawrence MC

EMDB-24927: 
Leg region of a complex of IGF-I with the ectodomain of a hybrid insulin receptor / type 1 insulin-like growth factor receptor
Method: single particle / : Xu Y, Lawrence MC

EMDB-13957: 
Extended H/L (SLPH/SLPL) complex from C. difficile (CD630 strain) fit into R20291 S-layer negative stain map
Method: electron crystallography / : Banerji O, Wilson JS, Bullough PA

PDB-7qgq: 
Extended H/L (SLPH/SLPL) complex from C. difficile (CD630 strain) fit into R20291 S-layer negative stain map
Method: electron crystallography / : Banerji O, Wilson JS, Bullough PA

EMDB-21415: 
Head region of the open conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II.
Method: single particle / : Xu Y, Kirk NS, Lawrence MC, Croll TI

EMDB-21416: 
Leg region of the open conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II.
Method: single particle / : Xu Y, Kirk NS, Lawrence MC, Croll TI

EMDB-21417: 
Head region of the closed conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II.
Method: single particle / : Xu Y, Kirk NS, Lawrence MC, Croll TI

EMDB-21418: 
Leg region of the closed conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II
Method: single particle / : Xu Y, Kirk NS, Lawrence MC, Croll TI

EMDB-0360: 
Saccharomyces cerevisiae spliceosomal E complex (UBC4)
Method: single particle / : Liu S, Li X

EMDB-0361: 
Saccharomyces cerevisiae spliceosomal E complex (ACT1)
Method: single particle / : Liu S, Li X

PDB-6n7p: 
S. cerevisiae spliceosomal E complex (UBC4)
Method: single particle / : Liu S, Li X, Zhou ZH, Zhao R

PDB-6n7r: 
Saccharomyces cerevisiae spliceosomal E complex (ACT1)
Method: single particle / : Liu S, Li X, Zhou ZH, Zhao R

PDB-6n7x: 
S. cerevisiae U1 snRNP
Method: single particle / : Li X, Liu S, Jiang J, Zhang L, Espinosa S, Hill RC, Hansen KC, Zhou ZH, Zhao R

EMDB-7109: 
S. cerevisiae spliceosomal post-catalytic P complex
Method: single particle / : Liu S, Li X, Zhou ZH, Zhao R

PDB-6bk8: 
S. cerevisiae spliceosomal post-catalytic P complex
Method: single particle / : Liu S, Li X, Zhou ZH, Zhao R

EMDB-8622: 
S. cerevisiae U1 snRNP
Method: single particle / : Li X, Liu S, Jiang J, Zhang L, Espinosa S, Hill RC, Hansen KC, Zhou ZH, Zhao R

EMDB-4016: 
A 4.5 Angstrom structure of HIV-1 CA-SP1
Method: subtomogram averaging / : Schur FKM, Obr M, Hagen WJH, Wan W, Arjen JJ, Kirkpatrick JM, Sachse C, Kraeusslich HG, Briggs JAG

EMDB-4017: 
A 4.2 Angstrom structure of HIV-1 CA-SP1 from immature HIV-1 particles
Method: subtomogram averaging / : Schur FKM, Obr M, Hagen WJH, Wan W, Arjen JJ, Kirkpatrick JM, Sachse C, Kraeusslich HG, Briggs JAG

EMDB-4018: 
Cryo-electron tomogram containing HIV-1 deltaMACANC virus-like particles assembled in vitro in presence of Bevirimat
Method: electron tomography / : Schur FKM, Obr M, Hagen WJH, Wan W, Arjen JJ, Kirkpatrick JM, Sachse C, Kraeusslich HG, Briggs JAG

EMDB-4019: 
Cryo-electron tomogram containing HIV-1 deltaMACANC virus-like particles assembled in vitro
Method: electron tomography / : Schur FKM, Obr M, Hagen WJH, Wan W, Arjen JJ, Kirkpatrick JM, Sachse C, Kraeusslich HG, Briggs JAG

EMDB-4020: 
Cryo-electron tomogram containing immature HIV-1 particles
Method: electron tomography / : Schur FKM, Obr M, Hagen WJH, Wan W, Arjen JJ, Kirkpatrick JM, Sachse C, Kraeusslich HG, Briggs JAG

EMDB-4015: 
A 3.9 Angstrom structure of HIV-1 CA-SP1 assembled in presence of Bevirimat
Method: subtomogram averaging / : Schur FKM, Obr M, Hagen WJH, Wan W, Arjen JJ, Kirkpatrick JM, Sachse C, Kraeusslich HG, Briggs JAG

PDB-5l93: 
An atomic model of HIV-1 CA-SP1 reveals structures regulating assembly and maturation
Method: subtomogram averaging / : Schur FKM, Obr M, Hagen WJH, Wan W, Arjen JJ, Kirkpatrick JM, Sachse C, Kraeusslich HG, Briggs JAG

EMDB-1484: 
Ribosome Binding of a Single Copy of the SecY Complex: Implications for Protein Translocation
Method: single particle / : Menetret JF, Schaletzky J, Clemons WM Jr, Osborne AR, Skanland SS, Denison C, Gygi SP, Kirkpatrick DS, Park E, Ludtke SJ, Rapoport TA, Akey CW

PDB-3bo0: 
Ribosome-SecY complex
Method: single particle / : Akey CW, Menetret JF

PDB-3bo1: 
Ribosome-SecY complex
Method: single particle / : Akey CW, Menetret JF
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
